Package script providing a complete processing chain for quality assessment of ALS flight strips.
opalsQuality calculates DSMs and visualizations thereof, point density maps, strip difference maps, and estimates of strip difference vectors.
The Python script opalsQuality is the main script for the corresponding opalsQuality package. It uses multiple sub-scripts to create different products for checking and documenting the quality of an ALS flight campaign. The following aspects are covered:
For estimating the actual quality of the captured ALS data, it is recommended to devide the area into parts considered for the derivation of quality measures and those, which should be exluded or masked out, replectively. Reasons for masking areas are manifold; examples are: rugged surface areas, areas covered by vegetation, scan shadows, water bodies, etc.
For controlling the masked areas, Python script opalsQuality contains the following parameters mask, gridMask and recalcMaskFiles, which are explained in detail in the following:
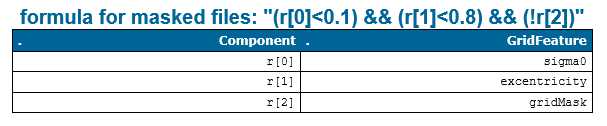
The mask parameter enables the user to create masked files using custom formulas for attributes calculated by opalsGrid, namely the opalsGridFeature. In the documentation of this module you can find the complete list of GridFeatures that can be calculated and later used as masks. The formulas should be given as Generic filter formulas. The syntax for specifying n formulas for n GridFeatures in the mask parameter should look like this.
The formulas are then combined and used in the computation. The standard formula for masked computations is specified at the top cfg file, which looks like this.
Parameter gridMask allows to specify a formula, which whether a negative mask (gridMask:"!r") is being used, for example to mask out unwanted areas like lakes or rivers. A positive mask (gridMask:"r") can be used to only compute certain areas of interests and mask out all areas which are not within the specified rasterfile.
As shown above implicitly, the script parameter mask can either be set in the commandline with the known syntax or as a paramater in the [script.opalsQuality]-section in the cfg-File.
Within the subscript *_grid*, the mask is combined and the final formula string is reported to the user via the opalsLog. Figure 1 shows an example of how the formula looks like with the above provided components.
The example 6 and the example 7 aim to show two uses cases for masking.
Finally, the boolean parameter recalcMaskFiles can be used to improve the masking in case the results of a previous run of opalsQuality call for an adaption of the masked areas. When activated in the repeat call of opalsQuality, this forces the re-creation of the mask files even if skipIfExists is set and the mask files (from the previous run) exist on disk.
Python script opalsQuality produces a series of intermediate and final results, which are stored in separate folders. The default folder structure is documented in the following tables:
| Default Directory | Produced by | Comments |
|---|---|---|
import | qltDSM, qltStripDiff, qltDensity, qltLSM, qltPrecision | The folder contains the imported ODM files. All sub scripts start with importing the data into the OPALS Datamanager format. |
grid | qltDSM, qltStripDiff, qltLSM, qltPrecision | The folder contains the DEM raster files for each strip of the flight block as well as additional grid feature raster files (sigma0, excentricity...) used for (i) masking and (ii) precision calculation. In addition, the folder conains the calculated mask for each strip and the masked/unmasked versions of the primary DEM and the sigma0 feature raster file. |
bounds | qltStripDiff | The folder contains the outlines of each strip (vector files contiaining the strip boundary polygon) as basis for the derivation of strip overlaps. |
cell | qltDensity | The folder contains the point density raster maps for each strip and the mosaic composed of all strips. |
diff | qltStripDiff | The folder contains the strip height difference raster maps for each strip pair and the mosaic composed of all strip pairs. If masking is activated, a separate raster is created for the masked and the unmasked version. |
overlap | qltStripDiff | The folder contains the file stripPairs.txt containing the list of overlapping strip pairs. |
| Default Directory | Produced by | Comments |
|---|---|---|
opalsQuality\Density | qltDensity | The folder contains sub-folders for the color-coded density maps (visu) and statistical point density analysis (histo). |
opalsQuality\DSM | qltDSM | The folder contains sub-folders for the hill shadings (shading) and color coded height maps (zcoding). |
opalsQuality\LSM | qltLSM | The folder contains the estimated transformation parameters for overlapping strip pairs in tabular form (LSM_TrafPars.csv) and as a parameter XML file (lsmOutput.xml). |
opalsQuality\Precision | qltPrecision | The folder contains sub-folders for masked and unmasked versions of the precision estimates (masked / unmasked). Each of the two folders consists of two sub-folders containing the color-coded height precision maps (zcoding) and the statistical analysis (histo). Separate files are generated for each strip and the mosaic composed of all strips. |
opalsQuality\StripDiff | qltStripDiff | As above, the folder contains sub-folders for masked and unmasked versions of the strip height differences. Each of the two folders consists of two sub-folders containing the color-coded height difference maps (zcoding) and the statistical analysis (histo). Separate files are generated for each strip pair and the mosaic composed of all strip pairs. |
| possible input | evaluates to |
|---|---|
| 1, true, yes, Boolean(True), True | Boolean(True) |
| 0, false, no, Boolean(False), False | Boolean(False) |
| possible input | evaluates to |
|---|---|
| 1, true, yes, Boolean(True), True | Boolean(True) |
| 0, false, no, Boolean(False), False | Boolean(False) |
| possible input | evaluates to |
|---|---|
| 1, true, yes, Boolean(True), True | Boolean(True) |
| 0, false, no, Boolean(False), False | Boolean(False) |
| possible input | evaluates to |
|---|---|
| 1, true, yes, Boolean(True), True | Boolean(True) |
| 0, false, no, Boolean(False), False | Boolean(False) |
The data used in the following examples can be found in the $OPALS_ROOT/demo/ directory.
In the first example, only the strip data file list is specified as input parameter:
striplist.txt may, e.g., contain the following lines:
Please note, that in the above example, strip31 and strip32 are ignored, since the corresponding line starts with a hash (=comment) character. Thus, the quality assessment is only carried out for the rest of the strips (strip11, strip21, strip21 and strip 22). Since no further parameters are specified, the output files are created in the current working directory ($OPALS_ROOT/demo/), all intermediate files are stored in subfolders of $OPALS_ROOT/demo/ and the default configuration files stored in $OPALS_ROOT/packages/cfg are used.
To use all strip*.laz files and store the resulting and temporary files each in a separate folders, type:
For examples 1 and 2 to work, it is necessary that the current working directory is $OPALS_ROOT/demo/. To achieve the same results from any current working directory, using the -p parameter, pointing to the directory containing the strip files, is recommended.
If only the strip differences should be calculated (disregarding already calculated intermediate outputs) for the overlapping strip pairs but not for the entire block, use the following command:
Please note, that parameter names can be abbreviated as long as they can be unique resolved. E.g. -m is too short since it matches -mosaic and -mask, but -mo is ok. Hence, the very same command as above can also be written as:
Text to come...
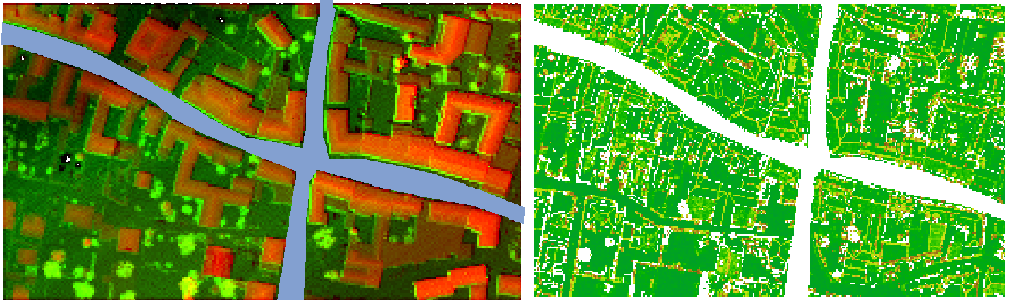
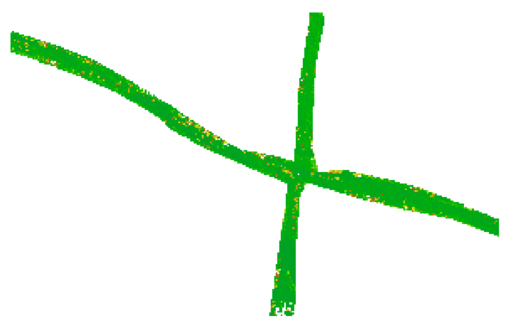
This first example should demonstrate the use of a negative mask. In this computation, the quality evaluation of the streets are not of interest, hence they are masked out. The raster file containing the approximate shape of the street is a rasterized shapefile called street_mask_ras.tif (see Figure 2a), which can be found in $OPALS_ROOT/demo/. Since a negative mask is desired, we do not need to change the default values in the cfg file. opalsQuality can therefore be called using the following command,
resulting in masked computations as shown in Figure 2b. It is clear to see that the masked out areas where excluded from the computation.
Following up on the hypothetical example of the street masks above: Let's say the streets are the only thing that is interesting for a certain application. In this case our mask file can be used as a positive mask to treat the street as an aoi. As only the masked files should be recaculated and all other files need not be computed again, the recalcMaskFiles parameter can be used to only compute files necessary for mask creation and reuse everything else. The opalsQuality call could therefore look like this:
Note the subtle change in the formula of GridMask within the mask parameter. Since "<" and ">" are reserved for redirections in the command shell, quotation signs are necessary to specify formulas that contain those symbols. The same can be achieved if the formula is changed within the cfg like so.
Then the call can look like this:
The result can be seen in Figure 3, now in the masked files only the street areas are taken into account.