The aim of Module Cell is to derive raster models by accumulating selected features (min, max, mean, etc.) of a selected input data attribute (z, amplitude, echo width, etc.).
Since ALS is a highly automated data capturing technique yielding an irregular point cloud, raster based data analysis is an important issue. Analyzing the data in regular raster structure is an established and convenient way to retrieve summary information about the data on a per cell basis. The following tasks can well be handled by raster analysis:
The main processing parameters of Module Cell are the list of features (parameter feature), for which raster models of a certain point attribute (parameter attribute) are to be derived. The OPALS Datamanager (ODM) is used as data base for deriving raster feature models. One of the outstanding properties of the ODM is that arbitrary point attributes (GPS time, amplitude, echo width ...) can be stored for each point along with the point coordinates (x,y,z). The default attribute is the point height (z), but raster feature models can also be generated for any of the available additional point attributes. For the derivation of a single raster pixel value, all points within the respective raster cell containing valid values for the selected attribute are queried from the ODM. The following cell operations (features) are supported:
The ODM administration of additional point attributes is highly extendable. Therefore, the list of possible attributes is only restricted by the availability of the respective information within the ODM. Section OPALS Datamanager contains a complete list of predefined ODM attributes.
The basic input for the raster model derivation is an ODM file (parameter inFile). Thus, the import of ALS point cloud data into the ODM is a prerequisite. Please refer to Module Import for more details concerning the import of ALS point cloud data. For each feature, the (aggregated) results are stored in separate raster files (parameter outFile) in GDAL supported data format (parameter oFormat). The raster derivation is organized in patches (tiles) internally, thus, maximum performance can be achieved for data formats supporting tile wise storage like GeoTiff or SCOP.RDH. The raster extents are either derived automatically based on the (xy-) extents of input data or can be specified by the user (parameter limit). Furthermore, the dimension of the raster cells (parameter cellSize) have to be specified. Optionally, a value indicating data voids (parameter noData) can be defined.
Beside raster files, also OPALS Data Managers (ODMs) may be generated as output files. In this case, the result of accumulation within each raster cell is not a single raster pixel value, but a single point. For the "selective" features (min, max, nmin, nmax, center), this is an original point with all its attributes; for the "aggregative" features (range, mean, median, rms, pdens, pcount, minority, majority, quantile, shannonEntropy, quadraticEntropy), this is a point just having 3 coordinates: x and y are given by the center of gravity of all points within the cell having a valid value for the selected attribute, whereas z is the respective attribute value as it is derived for a pixel in case of raster output files.
By default, the entire ODM contents are used as input data. However, in some cases it is useful to consider only a subset of the data, eg. to derive the point density for all last echoes. For this purpose, OPALS provides a general concept for filtering input data (parameter filter) by passing a Well-Known Text string (WKT). For a detailed description of the WKT filter syntax along with some illustrative examples, please refer to Section Filters.
The data set fullwave2.fwf used in the following examples is located in the $OPALS_ROOT/demo/ directory. The data set contains full waveform ALS point data (x, y, z, GPS time, amplitude, echo width, echo number, echo qualifier) of a partially wooded area. This module requires an OPALS data manager file as input, which can be generated by using Module Import as follows:
Note: The following figures have been generated by applying Module ZColor to the output files (using the default color palette).
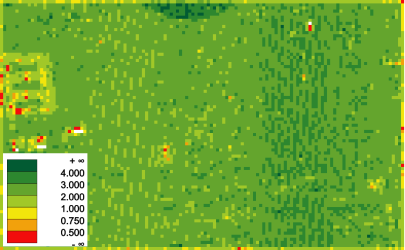
In this example, a point density map is produced (output: pdens.tif).
The file format (GTiff) is estimated from the file extension. The feature pdens (point density) is specified and quadratic cells with a size of 2.0 m are used for accumulation. In this case, the selected attribute is not relevant, because just the number of points per cell (divided by the cell area) determines the result. The extents are determined from the bounding box of the entire ODM point data set.

Since all echoes are considered, the regions with multiple echoes (i.e.
vegetation) show a higher point density.
In this example, two raster models are derived from the amplitude values of the first echoes for the features min and max (output: amplFirst_min.tif and amplFirst_max.tif).
The file format (GTiff) is estimated from the file extension. The two output files names are based on the specified outFile and the features. As attribute the amplitude is selected. A filter is specified in such a way that only first echoes are considered. Quadratic cells with a size of 2.0 m are used for accumulation. The extents are determined from the bounding box of the entire ODM point data set.
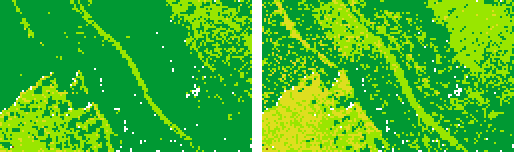
As expected, in vegetated areas, the first-echo-amplitudes tend to be smaller.
In this example, two raster models are derived from the echo width values of the first/last echoes for the feature max (output: maxEchoWidthFirst.tif and maxEchoWidthLast.tif).
The file format (GTiff) is estimated from the file extension. As attribute the echo width is selected. Filters are specified to select (i) only first echoes, (ii) only last echoes, or (iii) only echoes with a width greater than 4.5ns. Please note that quoting the filter string is necessary in the latter case due to the embedded blanks and special characters. Quadratic cells with a size of 2.0 m are used for accumulation. The extents are determined from the bounding box of the entire ODM point data set.

Comparing the two models, one can clearly see that high echo widths are occurring rather at first echoes than at last echoes.
This examples illustrates the multiple input ODM file capabilities of module Cell using the demo data strip*1.laz. As a prerequisite, import the data files into separate OPALS data managers using the following commands:
To compute the point density of an entire flight block multiple input ODM files can be specified in a single program call. A colourized version of the resulting point density map is illustrated in the below figure: