- See also
- python.workflows.forDelineation
Aim of script
determine the forest boundary from topographic models (namely an nDSM, a DTM and any kind of roughness indicator map) derived from 3D point clouds e.g. ALS data or point clouds from image matching
General description
This python scripts attempts to delineate forest areas from topographic models derived from ALS Data or image matching. Definitions of forests vary widely between countries and are difficult to express in mathematical (i.e. deterministic) terms. In many forest definitions, crown coverage (area covered by tree crowns/reference area) plays a crucial role, but definitions of the reference area are often missing. Eysn et. al., 2012 use the convex hull of a triplet of trees (determined by calculating a Delaunay triangulation on the tree positions in a forest) as a reference area. Other criteria in forest definitions include minimum (normalized) height, minimum area and width.
As the crown coverage approach from Eysn et. al., 2012 is implemented in forDelineation single tree detection is an important. Single trees are extracted with the forTreeDetection.py - script. It is a raster-based approach using the nDSM as an input. The focus of this single tree detection approach is to extract single trees for areas with low crown coverage (i.e. along the timber line) with high completeness. To avoid trees on roofs a differentiation between roofs and forests is needed. This is done based on a surface roughness layer. This may be a slope adaped EchoRatio Map (created using Module EchoRatio), but can also be e.g. a sigma0 map resulting as a by-product from DSM calculation (using Module Grid with the -feat sigma0 flag). Depending on the choice of roughness map, rougher areas can have higher or lower values than smooth areas, therefore caution has to be taken when setting the threshold formula (see Parameter description).
Forest delineation is difficult in areas with young trees, since height, roughness and crown coverage criteria might not be met, but it should clearly be included in the forest areas. Other problems arise in corn fields (corn can grow up to similar heights than those of young forests and meet the required criteria) and in suburban regions with trees and small buildings, where a forest area might be detected, having only three trees and a house in-between (appearing as a crown in the nDSM). Finally, snow can has an impact on the nDSM and the surface roughness of young forests, which can lead to incorrectly delineated forests.
Basic algorithm
The algorithm consists the following steps:
- Binarising the roughness map using the provided threshold formula.
- Using morphological operations (close) to remove borders of houses and power lines (esp. in sER maps)
- Running forTreeDetection to detect trees
- Selecting isolated, free-standing trees for a calibration of a formula that calculates crown radii from DTM height and tree(=nDSM) height at the positions of the isolated trees
- Triangulating the tree positions - for all detected trees
- Calculation of crown coverage based on the coefficients of the calibrated formula and the tree triangulation - removal of triangles that don't fit the criterea
- Buffering the triangles to ensure that the whole crown is represented, and not only the stems (by intersecting with the sER map).
- Applying minimum area (by vectorising) and minimum width (by using morphological operations) critera
- Repeat the last two steps for finalisation and smooth the derived model by morphologic operations (i.e. opening and then closing).
Usage of script
This script implements the preTilingManager framework to cope with large areas. This means, input can be pre-tiled, with or without overlap, but can also be one file per class (dsm, er, dtm). Proper overlap during the tiling will be ensured (default and minimum: -minArea/-minWidth.
Three (classes of) input files have to be provided: a DSM (-dsm), a DTM (-dtm) and a roughness map (e.g. EchoRatio, -echoRatio). All other parameters are optional. The files can be provided as absoulte or relative paths to single files or with wildcards (e.g. "DTMs\*.tif"). These files have to have a compatible grid layout, e.g. a linear combination of a corner of a pixel and the pixel size of any file must be able to get the corner of a pixel in the other input files. It is possible to run the opals.tools.rasterfun.resample_grid function to achieve this (for input files with the same boundaries).
Parameter description
-dsm DSM File
Type : Path
Remark : mandatory
-ndsm nDSM File
Type : Path
Remark : optional
Description: If no nDSM is provided it will be calculated
-echoRatio slope adaptive echoratio map (or other)
Type : Path
Remark : mandatory
-dtm DTM File
Type : Path
Remark : mandatory
-workingDir working directory
Type : Path
Remark : optional, default: current directory
Description: working directory - All results will be stored here (folders will be generated).
-minHeight minimum tree height for forested areas [m].
Type : Floating-point number
Remark : optional, default: 2
-percentageCrownCoverage minimum threshold of crown coverage for forested areas [%%]
Type : Floating-point number
Remark : optional, default: 30
-triangleLengthCrownCoverage maximum threshold for triangle side length [m]
Type : Floating-point number
Remark : optional, default: 20
-minArea minimum area for forested areas [m^2]
Type : Floating-point number
Remark : optional, default: 500
-minWidth minimum width for forested areas [m]
Type : Floating-point number
Remark : optional, default: 10
-truncMaxHeight upper limit for height values (nDSM,DTM) [m]
Type : Floating-point number
Remark : optional, default: (45, 3000)
Description: Upper threshold for truncating too high maximas and high areas above the upper timberline. The first value indicates the treshold for the nDSM. The second value indicates the treshold for the DTM. Example: (50,2100) (in meters).
-tiling tiling for processing and output
Type : String
Remark : optional
Description: Set tiling for processing and output. Can be one of [ndsm, dtm, er] to use the original tiling of the respective input files. Can also be an integer representing a side length (in meters), or a path to a shapefile containing tiling polygons (overlap will be added for calculation).
-naming naming convention for tiled files
Type : String
Remark : optional, default: LL_6
Description: Set naming convention for tiled files (eg. a corner + delimiter + nDigits --> "LL_4", or a shapefile column name when using shapefiles, or NUMBERED or INDEX).
-buffer buffer size for tiling
Type : Floating-point number
Remark : optional
Description: Override automatic calculation of buffer size for tiling. (Can only be bigger than calculated buffer=minarea/minwidth
-removeTempFiles remove temporary files
Type : Boolean
Remark : optional, default: False
Description: Remove the temporary files for each tile after calculation (saves disk space)
| possible input | evaluates to |
| 1, true, yes, Boolean(True), True | Boolean(True) |
| 0, false, no, Boolean(False), False | Boolean(False) |
-skipIfExists skip tile if result is in target directory
Type : Integer
Remark : optional, default: 0
Description: Skip processing of a tile if the result file(s) already exist in the target directory: 0=Do not skip, 1=Skip tile.
-mosaic Output a mosaic file covering the whole area of interest
Type : Boolean
Remark : optional, default: True
| possible input | evaluates to |
| 1, true, yes, Boolean(True), True | Boolean(True) |
| 0, false, no, Boolean(False), False | Boolean(False) |
-regressionParameters regression paramters j, k, l
Type : Floating-point number
Remark : optional, default: (0.85462, 0.06511, 0.00045234)
-edgeCorrectionFormula forumla to correct crowns on the forest edge
Type : String
Remark : optional, default: r[0] < 85
Description: Formula used on forest edges to regain crowns that were cut off in the triangulation process. Same nomenclature as -thresholdFormulaEchoRatio.
-distribute The number of processes created by preCutting. Use 0 for maximum number of threads.
Type : Integer
Remark : optional, default: 1
Relation tree height vs. crown diameter (based on NFI Data)
Relation formula: crown diameter = j + k * treeheight + l * elevation
-calibCrowns calibrate a function for crown diameter calculation
Type : Boolean
Remark : optional, default: True
Description: Calibrate a function for estimating the tree crowns based on ALS data. If set to FALSE no calibration will be carried out. The specified values of the flags j, k and l are used then.
| possible input | evaluates to |
| 1, true, yes, Boolean(True), True | Boolean(True) |
| 0, false, no, Boolean(False), False | Boolean(False) |
-thresholdFormulaEchoRatio forumla for sER threshold
Type : String
Remark : optional, default: r[0] < 85
Description: Formula for thresholding sER (or other) map for further enhancements, with r[0] representing the sER (or other) map, r[1] representing the DTM and r[2] representing the ndsm. Should return True for forest areas.
-morphEchoRatio kernelsize for opalsmorph
Type : Floating-point number
Remark : optional, default: 2
Description: Kernelsize for opalsmorph for enhancing sER (or other) map (removement of building edges etc.)
Logging Options
Settings concerning the verbosity level of logging.
-fileLogLevel Log level in the logfile
Type : LogLevel
Remark : optional, default: info
-screenLogLevel Log level on screen
Type : LogLevel
Remark : optional, default: info
-logger Logger
Type : Logger
Remark : optional
Description: Logger is usually provided by the opals framework.The user may provide their own logger object, but it has to function in the same way as the opals Logger.
Examples
The data used in the following examples can be found in the $OPALS_ROOT/demo/forDelineation directory.
Example 1
This assumes three files in the Test_Data directory, they are not distributed with opals. It will create a "project_dir" for the results and use a minimum width of 6 meters, a minimum crown coverage of 10%, tile the input files into two columns and two rows (resulting in four tiles), and overwrite existing files (instead of skipping calculation)
opals forDelineation -dsm forDelineation\DSM\DSM.tif -echoRatio forDelineation\ER\ER.tif -dtm forDelineation\DTM\DTM.tif -
minHeight 6 -percentageCrownCoverage 10 -
tiling 2r2c -skipIfExists 0

Figure 1: The result of the forest delineation for one tile. White pixels represents forest areas.
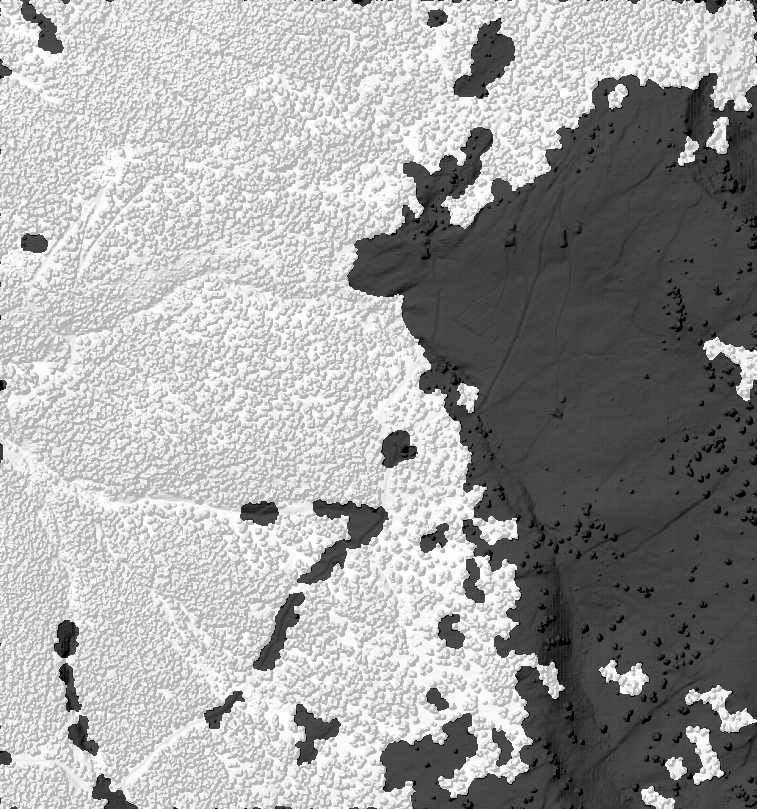
Figure 2: If -mosaic is true a mosaic of all tiles will also be created. In the background is the hillshaded DSM file for reference.
Example 2
This runs on directories containing multiple input files, counts areas above 1000m and with an nDSM values higher than 3m as forest (both for setting the threshold and for buffering the result)
opals forDelineation -dsm forDelineation\DSM\*.tif -echoRatio forDelineation\ER\*.tif -dtm forDelineation\DTM\*.tif -thresholdFormulaEchoRatio
"r[0] < 85 and r[1] > 1000 and r[2] > 3" -
tiling 2r2c -skipIfExists 0 -edgeCorrectionFormula
"r[0] < 85 and r[1] > 1000 and r[2] > 3"
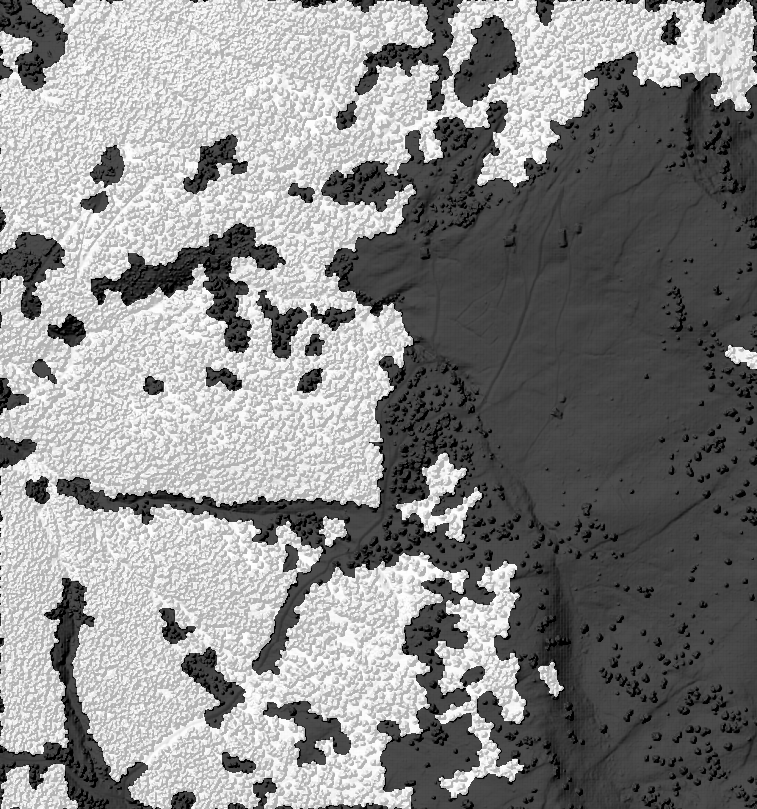
Figure 3: The resulting mosaic with different formulas and parameters. In the background is the hillshaded DSM file for reference.
Example 3
This example shows that it is possible to use algebraic expressions in the forest definition, e.g. the higher the elevation, the lower the tree height may be. No tiling is applied here, therefore one single tile covering the whole region will be created:
opals forDelineation -dsm forDelineation\DSM\*.tif -echoRatio forDelineation\ER\*.tif -dtm forDelineation\DTM\*.tif -thresholdFormulaEchoRatio
"r[0] < 85 and r[2] > 3000/r[1]" -edgeCorrectionFormula
"r[0] < 85 and r[2] > 3000/r[1]"

Figure 4: The resulting mosaic with different formulas and parameters. In the background is the hillshaded DSM file for reference.



